Ivana Borovská, Chundan Zhang, Sarah-Luisa J. Dülk, Edoardo Morandi, Marta Cardoso, Daphne A.L. van den Homberg, Michael T. Wolfinger, Willem A. Velema, and Danny Incarnato
RNA molecules can populate ensembles of alternative structural conformations, but comprehensively mapping RNA conformational landscapes within
living cells presents significant challenges and has, as such, so far remained elusive. Here, we generate transcriptome-scale maps of RNA secondary
structure ensembles in both Escherichia coli and human cells, uncovering features of structurally-heterogeneous regions. By combining ensemble
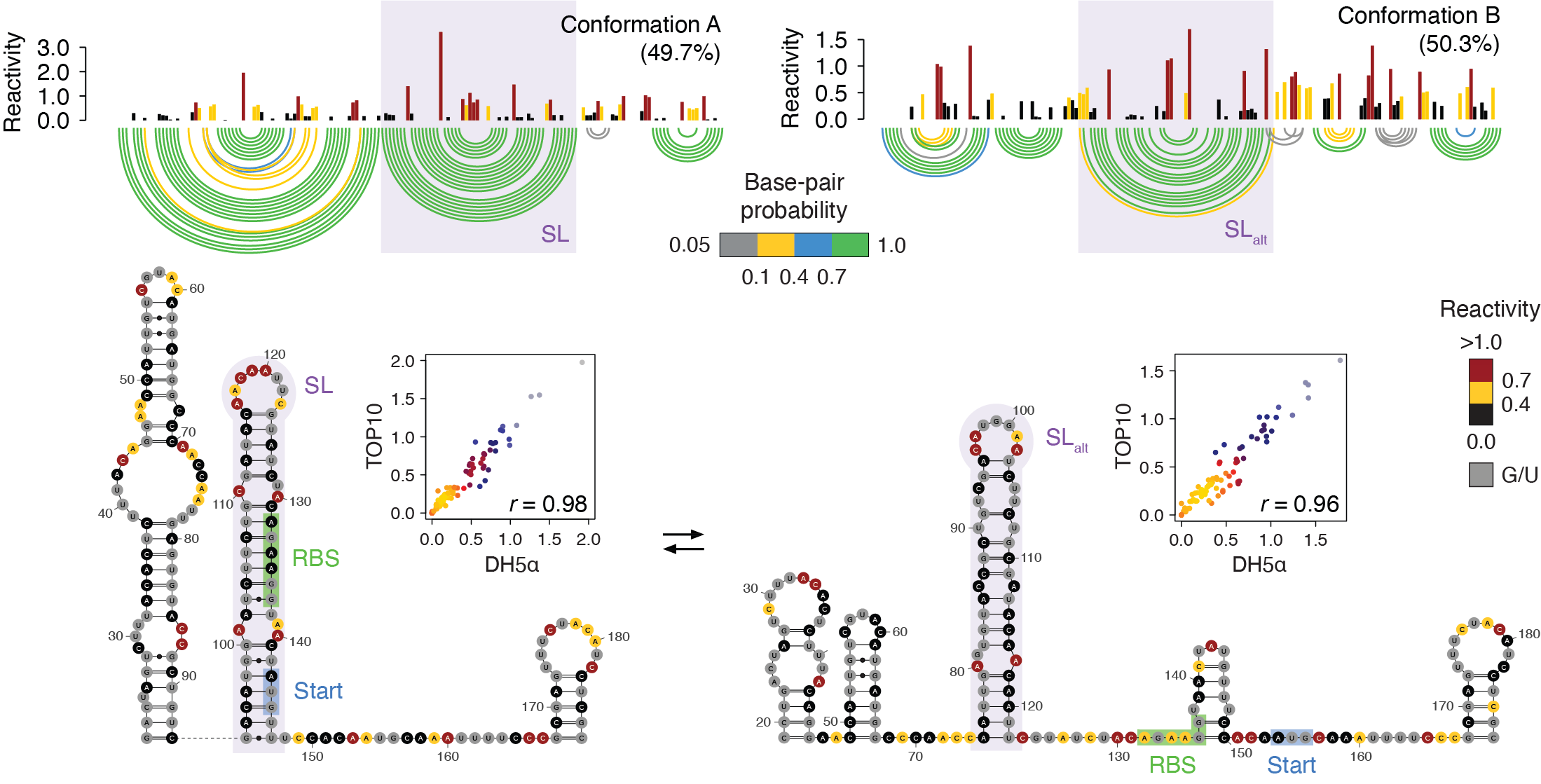
deconvolution and covariation analyses, we report the discovery of several bacterial RNA thermometers in the 5′ UTRs of the cspG, cspI,
cpxP and lpxP mRNAs of E. coli. We mechanistically characterize how these thermometers switch structure in response to cold shock
and revealed the CspE chaperone-mediated regulation of lpxP. Furthermore, we introduce a method for the transcriptome-scale mapping of 5′ UTR
structures in eukaryotes and leverage it to uncover RNA structural switches regulating the differential usage of open reading frames in the 5′ UTRs of
the CKS2 and TXNL4A mRNAs in HEK293 cells. Collectively, this work reveals the complexity of RNA structural dynamics in living cells, and
provides a resource to accelerate the discovery of regulatory RNA switches.
| E. coli transcriptional unit annotation | Download | |
| E. coli, RC files (RNA Framework-compliant) | Download | |
| E. coli, 37°C, Normalized reactivity XML files (RNA Framework-compliant) | Download | |
| E. coli, 37°C, Structure models | Download | |
| E. coli, 37°C, Conserved structures | Browse | |
| E. coli, 10°C, Normalized reactivity XML files (RNA Framework-compliant) | Download | |
| E. coli, 10°C, Structure models | Download | |
| HEK293, RC files (RNA Framework-compliant) | Download | |
| MM files (DRACO-compliant) | Zenodo | |
| Stockholm alignments | Download | |
| DeConStruct pipeline | GitHub |