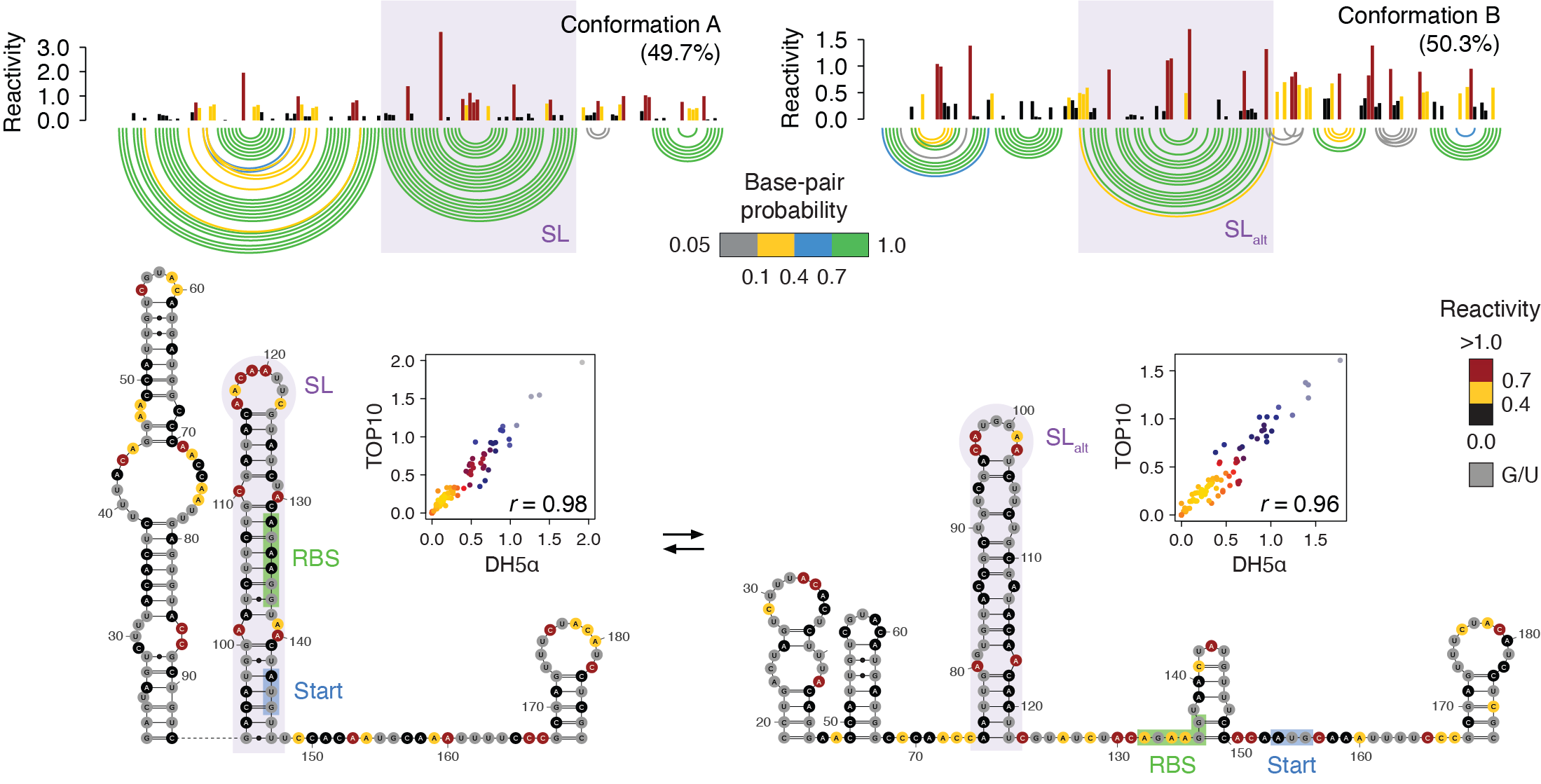
Identification of conserved RNA regulatory switches in living cells using RNA secondary structure ensemble mapping and covariation analysis
Structural and biophysical dissection of RNA conformational ensembles
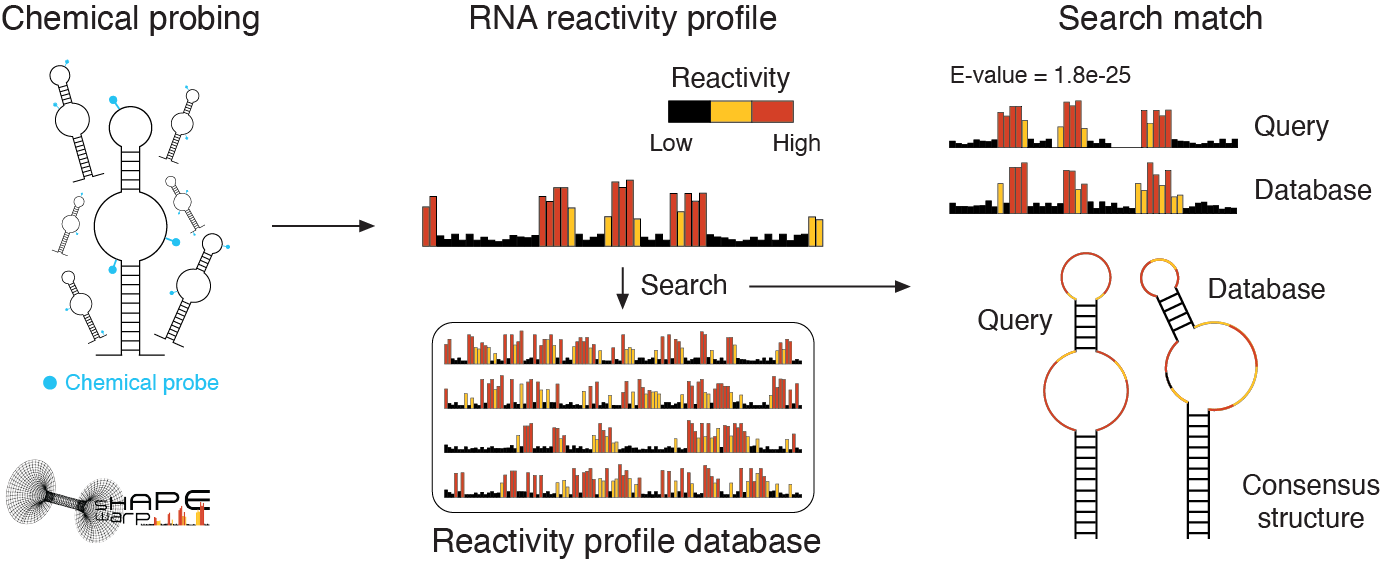
SHAPEwarp-web: sequence-agnostic search for structurally homologous RNA regions across databases of chemical probing data
Probing the dynamic RNA structurome and its functions
SHAPE-guided RNA structure homology search and motif discovery
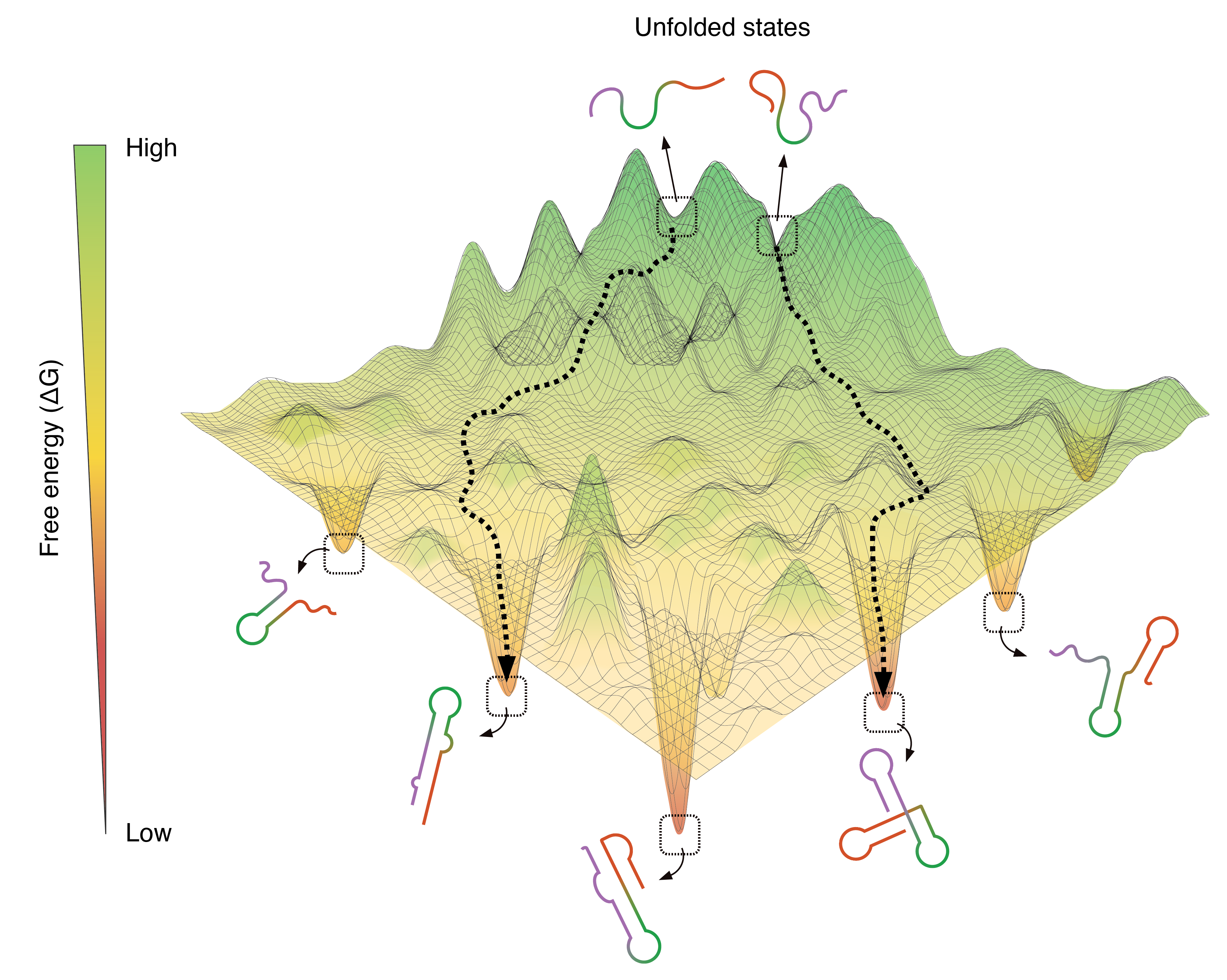
Genome-scale deconvolution of RNA structure ensembles
A novel SHAPE reagent enables the analysis of RNA structure in living cells with unprecedented accuracy
Structure and regulation of coronavirus genomes: state-of-the-art and novel insights from SARS-CoV-2 studies
Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements
In vivo analysis of influenza A mRNA secondary structures identifies critical regulatory motifs
RNA Framework: an all-in-one toolkit for the analysis of RNA structures and post-transcriptional modifications
The RNA Epistructurome: Uncovering RNA Function by Studying Structure and Post-Transcriptional Modifications
In vivo probing of nascent RNA structures reveals principles of cotranscriptional folding
High-throughput single-base resolution mapping of RNA 2'-O-methylated residues
Methylation-assisted bisulfite sequencing to simultaneously map 5fC and 5caC on a genome-wide scale for DNA demethylation analysis
RNA structure framework: automated transcriptome-wide reconstruction of RNA secondary structures from high-throughput structure probing data
Single-Base Resolution Analysis of 5-Formyl and 5-Carboxyl Cytosine Reveals Promoter DNA Methylation Dynamics
Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome
MREdictor: a two-step dynamic interaction model that accounts for mRNA accessibility and Pumilio binding accurately predicts microRNA targets