Lisa Marie Simon, Edoardo Morandi, Anna Luganini, Giorgio Gribaudo, Luis Martinez-Sobrido, Douglas H. Turner, Salvatore Oliviero and Danny Incarnato
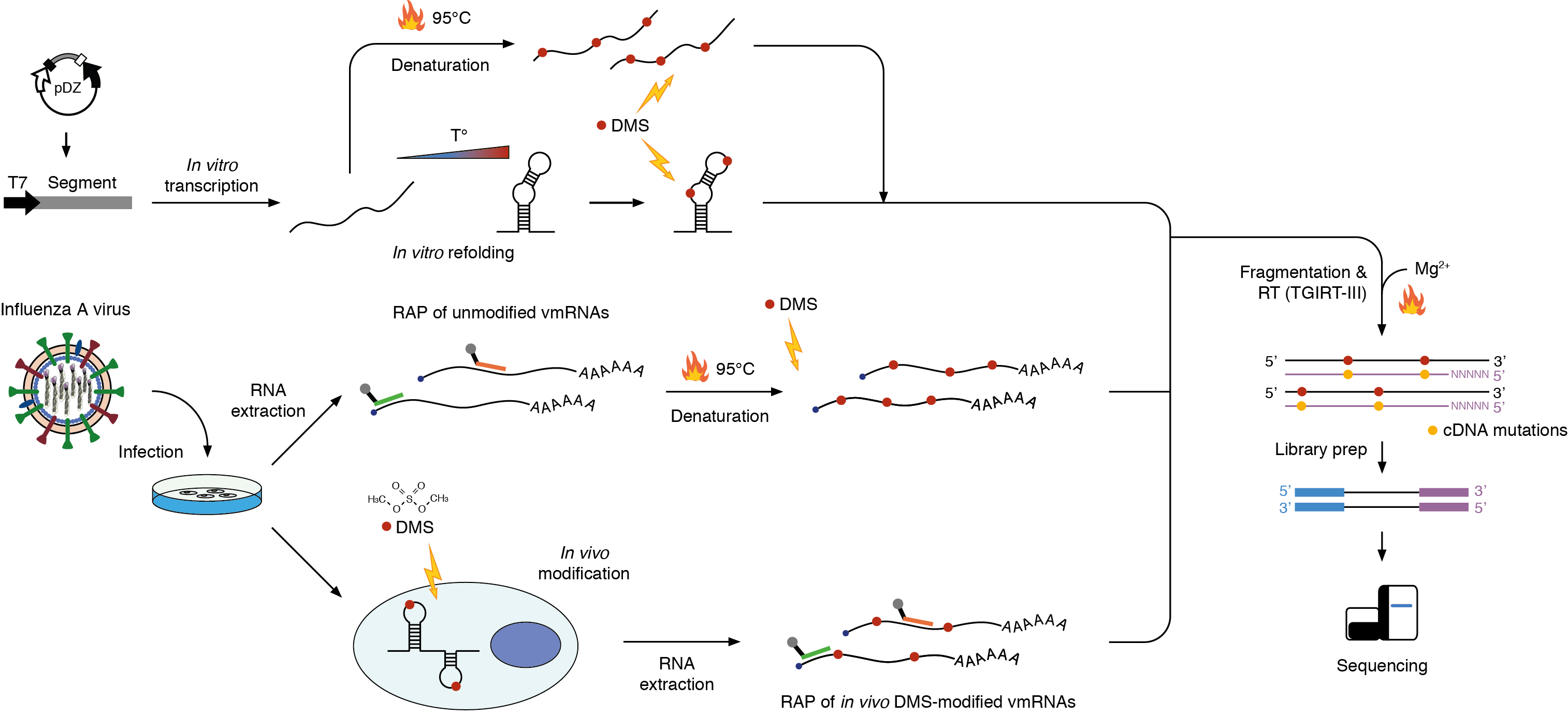
The influenza A virus (IAV) is a continuous health threat to humans as well as animals due to its recurring epidemics and pandemics. The IAV genome is segmented and the eight negative-sense viral RNAs (vRNAs) are transcribed into positive sense complementary RNAs (cRNAs) and viral messenger RNAs (vmRNAs) inside infected host cells. A role for the secondary structure of vmRNAs has been hypothesized and debated for many years, but knowledge on the structure vmRNAs adopt in vivo is currently missing. Here we solve, for the first time, the in vivo secondary structure of IAV vmRNAs in living infected cells. We demonstrate that, compared to the in vitro refolded structure, in vivo vmRNAs are less structured but exhibit specific locally stable elements. Moreover, we show that the targeted disruption of these high-confidence structured domains results in an extraordinary attenuation of IAV replicative capacity. Collectively, our data provide the first comprehensive map of the in vivo structural landscape of IAV vmRNAs, hence providing the means for the development of new RNA-targeted antivirals.