Edoardo Morandi, Martijn J. van Hemert and Danny Incarnato
The rapidly growing popularity of RNA structure probing methods is leading to increasingly large amounts of available RNA structure information.
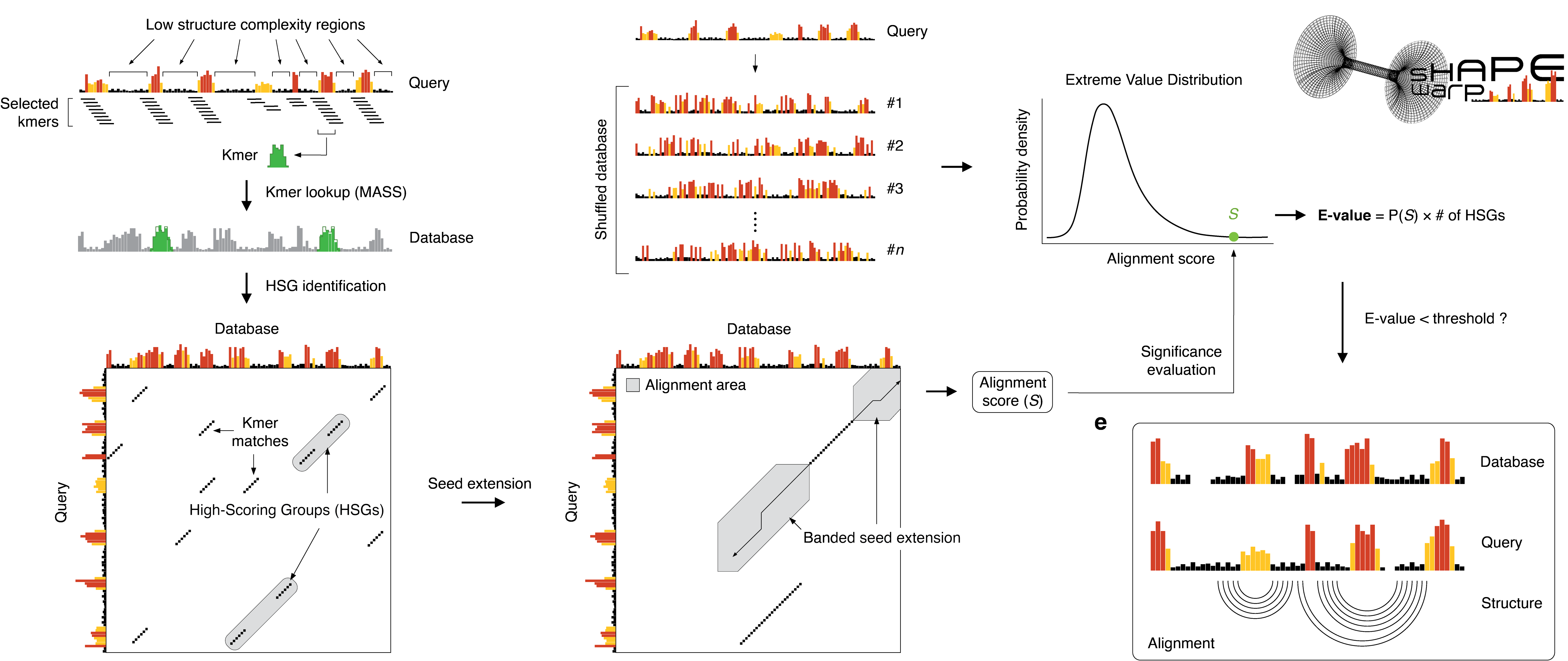
This demands the development of efficient tools for the identification of RNAs sharing regions of structural similarity by direct comparison of
their reactivity profiles, hence enabling the discovery of conserved structural features. We here introduce SHAPEwarp, a largely sequence-agnostic
SHAPE-guided algorithm for the identification of structurally-similar regions in RNA molecules. Analysis of Dengue, Zika and coronavirus genomes
recapitulates known regulatory RNA structures and identifies novel highly-conserved structural elements. This work represents the first step towards
the model-free search and identification of shared RNA structural features within transcriptomes.
| SHAPE data (RNA Framework-compliant XML files) | Download | |
| SHAPEwarp databases | Download | |
| SHAPEwarp queries | Download | |
| Alignments for elements identified by SHAPEwarp+cm-builder (Stockholm files) | Download | |
| Covariance models for elements identified by SHAPEwarp+cm-builder | Download | |
| SHAPEwarp algorithm (source code) | GitHub |